Biochemists find incomplete protein digestion is a useful thing for some bacteria
(Phys.org) —Usually indigestion is a bad thing, but experiments by researcher Peter Chien and graduate student Robert Vass at the University of Massachusetts Amherst recently showed that for the bacteria Caulobacter crescentus, partial degradation of a DNA replication protein is required to keep it alive.
DNA replication is one of the most highly controlled biological processes in all organisms, says Chien, an assistant professor of biochemistry and molecular biology at UMass Amherst. From humans all the way back to bacteria, all cells must faithfully duplicate their genomes in order to survive. To coordinate the start, ensure the completion and repair damages during DNA replication, specialized proteins play a key role by regulating processes.
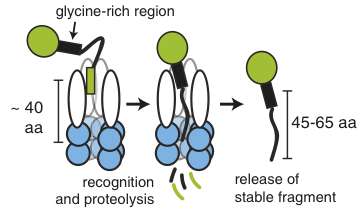
In work published this month in Proceedings of the National Academy of Sciences, Chien and Vass report that one of these specialized replication factors, DnaX, is, to their surprise, partially digested or trimmed, physically cut into shorter fragments, by an energy-dependent protease known as ClpXP, which generates specific-sized fragments that are essential for Caulobacter's normal growth.
The phenomenon isn't entirely unknown, Chien explains. Short as well as long versions of DnaX had been observed 20 years ago in another bacteria, E. coli. But in E. coli, the short form was produced by changes in translation due to an early ribosome stop at a specific RNA sequence. That RNA sequence is absent in many bacteria DnaX genes including in Caulobacter, so scientists long thought that short DnaX only existed in bacteria like E. coli.
Based on previous studies in his lab, Chien and Vass moved on to investigate, by purifying these proteins and testing their activity in Caulobacter in vivo, whether the protein DnaX could be degraded by the protease ClpXP. To their surprise, ClpXP did recognize DnaX, but only partially degraded it, generating stable shorter fragments both in vitro and in vivo.
Further, they found that both long and short forms were essential for normal growth and that the short form was particularly important for DNA damage tolerance, suggesting that these fragments are not accidental but are made in a programmed manner.
Vass and Chien's new work shows that in some bacteria, short forms of DnaX are made instead by another method, proteolysis, with dramatic consequences for the cell, the biochemist says. "This type of convergent evolution results in similar protein outcomes, that is the generation of two forms through radically different mechanisms. This suggests that both long and short forms are crucial for all bacteria," he adds.
Protein degradation by energy-dependent proteases normally results in the complete destruction of target proteins, Chien notes. However, under particularly harsh artificial conditions in the test tube, these proteases can stall on certain targets. But until the recent UMass Amherst experiments, such an effect had never been seen inside a living bacterial cell, he adds.
Chien feels this mode of partial digestion could be useful in other bacteria to generate new functions from existing proteins and increasing the ability of a single gene to encode for multiple protein functions. This work was supported by NIH's National Institute for General Medical Sciences and by UMass Amherst.
More information: www.pnas.org/content/early/201 … /1311302110.abstract
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Massachusetts Amherst