New web tool simplifies prediction of regulatory networks in bacteria
From the lab to the computer: Scientists from the University of Freiburg have developed a computer program to predict the functions of bacterial gene regulators. This online software which is called CopraRNA could save researchers a lot of wet lab work as it precisely predicts which bacterial genes are controlled by certain regulators.
Scientists in the research group for Genetics and Experimental Bioinformatics led by Prof Dr Wolfgang Hess and the research group for Bioinformatics led by Prof Dr Rolf Backofen, a Member of the Cluster of Excellence BIOSS Centre for Biological Signalling Studies, both from the University of Freiburg, cooperated with a research group headed by Prof Dr Jörg Vogel from the University of Würzburg to compare regulatory mechanisms of related bacteria. This comparison is the basis of the computer program CopraRNA. CopraRNA reduces complicated laboratory tests while simplifying the search for bacterial regulators. It has been published in the scientific journal Proceedings of the National Academy of Sciences. Understanding the functions of these regulatory molecules is vital in order to fight pathogens in medicine. Furthermore their modification may aid in biotechnological projects.
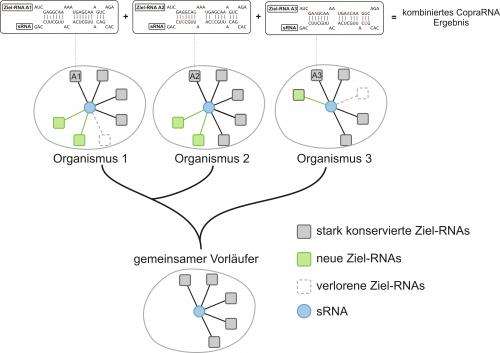
Unlike most molecules that control cell, the regulators investigated in this study are not proteins; they are RNA molecules. These ribonucleic acids, like DNA, consist of a code of four nucleobases, which form a chain connected by a sugar phosphate backbone. In the past, RNAs were only regarded as working copies of DNA for protein synthesis. The RNA molecules studied here do not produce proteins, however. For this reason, they are called non-coding RNA molecules, or ncRNAs. It was recently discovered that these molecules play a key role in the signalling network of a cell - for example, when a bacterium reacts to external stimulants. As a regulator, the RNA binds to many different targets in the bacterial cell, thereby regulating protein synthesis.
Thousands of newly discovered ncRNAs act as regulators in bacterial cellular systems. Researching their function in the laboratory requires much effort. Many comprehensive tests are necessary to determine which genetic sequences they interact with. We can simplify this process by predicting which genes these molecules might target for activation or inactivation. Both are based on the individual sequences of the ncRNA regulators. That is why the scientists have developed the CopraRNA software program as part of the collaborative project "ebio: RNAsys - Systembiologie der RNA", which is funded by the German Ministry of Education and Research. Researchers can now enter the RNA sequence of three or more bacterial species on the website rna.informatik.uni-freiburg.de/CopraRNA/ to acquire information about their possible functions. Whether or not the prediction is correct must then be followed up by experiments in the lab.
More information: Wright, P.R., et al. (in press). Comparative genomics boosts target prediction for bacterial small RNAs, Proc Natl Acad Sci. August 26, 2013, Early Edition. DOI: 10.1073/pnas.1303248110
Journal information: Proceedings of the National Academy of Sciences
Provided by Albert Ludwigs University of Freiburg