'Insect soup' holds DNA key for monitoring biodiversity

Scientists at the University of East Anglia have shown that sequencing the DNA of crushed up creepy crawlies can accelerate the monitoring and cataloguing of biodiversity around the world.
Research published today in the journal Ecology Letters shows that a process known as 'metabarcoding' is much faster than and just as reliable as standard biodiversity datasets assembled with traditional labour-intensive methods.
The breakthrough means that changing environments and endangered species can be monitored more easily than ever before. It could help researchers find endangered tree kangaroos in Papua New Guinea, discover which moths will be wiped out by climate change, and restore nature to heathlands in the UK, rubber plantations in China, and oil-palm plantations in Sumatra.
Lead researcher Dr Douglas Yu, from UEA's school of Biological Sciences, said: "Every living organism contains DNA, and even small fragments of that DNA can be used to identify species.
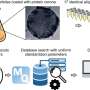
"We collected lots of insects and other creepy-crawlies, ground them up into an 'insect soup', and read the DNA using sequencers that are now cheap enough to use weekly or even daily.
"We compared our results with high-quality datasets collected in Malaysia, China and the UK which combined more than 55,000 arthropod and bird specimens and took experts 2,505 hours to identify. These kinds of datasets are the gold standard for biodiversity monitoring but are so expensive to compile that that we cannot use them for regular monitoring. Thus, conservation biologists and environmental managers are forced to work with little information.
"We found that our 'soup' samples give us the same biodiversity information as the gold-standard datasets. They are also more comprehensive, many times quicker to produce, less reliant on taxonomic expertise, and they have the added advantage of being verifiable by third parties."
The findings are important because they show that metabarcoding can be used to reliably inform policy and environmental management decisions.
Dr Yu added: "If the environment changes for the better or for the worse, what lives in that environment changes as well. Insect soup becomes a sensitive thermometer for the state of nature.
"For instance, we showed that if the UK Forestry Commission ploughs up some of the grass-covered trackways that run between our endangered heathland habitats, populations of rare spiders, beetles, and other creepy-crawlies can reconnect along those trackways, helping to stave off extinction.
"We are now working with the WWF and Copenhagen University to apply the method to bloodsucking leeches to look for endangered mammals in Vietnamese and Laotian rainforests. By creating a 'leech soup' we can get a list of mammals and know more about whether park conservation is working."
Each soup combines hundreds to thousands of insects caught using insect traps. The numbers captured amount to a tiny fraction of their overall populations and pose no threat to endangered species.
More information: doi.wiley.com/10.1111/ele.12162
Journal information: Ecology Letters
Provided by University of East Anglia










.jpg)




