Subset of short genes hidden inside plant genomes may be important in setting plant growth patterns

Although thousands of entire genomes have been sequenced, our understanding of their detailed workings remains far from complete. Researchers continue to find new genes, determine their function, and map how they interact to build organisms. Working on the well-studied model plant Arabidopsis thaliana, Kousuke Hanada and colleagues from the RIKEN Plant Science Center have revealed that a subset of tiny genes scattered through the genome may control the patterning of development.
To discover new genes, researchers must first mine huge volumes of genomic data to locate sections that have the basic structure of a gene, called 'open reading frames' (ORFs). Hanada's team focused on short ORFs (sORFs), which encode sequences of just 30–100 amino acids—compared with the average gene length of 600 amino acids in A. thaliana. Such sequences are often overlooked during annotation of the genome.
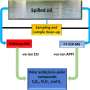
The research team identified nearly 8,000 sORFs through computer analysis of the A. thaliana genome. They then used expression analysis to exclude pseudogenes—genetic remnants that are no longer switched on or 'expressed'. The team achieved this by using a specially designed fluorescence microarray to generate an 'expression atlas' of highly expressed—and therefore likely to be functional—genes. On the microarray chip, genes fluoresce with an intensity proportional to their expression level. Through this approach, the researchers showed that 27% of the originally identified sORFs were highly expressed.
Hanada and colleagues then compared these likely functional sequences with those in 16 other plant genomes to identify which of their sORFs correspond to genes present in multiple species and are therefore highly conserved. This comparative genomics approach makes it possible to identify genes that have been evolutionarily conserved due to the essential functions they provide. The search revealed that more than half of the sORFs were similar to genes from other species.
Taking the 473 highly expressed and highly conserved sORFs, the researchers produced mutant A. thaliana plants in which these genes were overexpressed. A surprisingly high proportion of the mutants, close to 10%, had unusual traits, such as altered size, pale leaves, bent stems, or flowers with five instead of four petals. In contrast, overexpression of the previously known functional genes for A. thaliana only produced visible alterations in 1.4% of mutants.
The research demonstrates that sORFs potentially have pronounced effects on plant growth and form, and highlights the importance of these tiny elements in the genome. "Future work will focus on determining how the genes function when expressed normally," says Hanada.
More information: Hanada, K. et al. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proceedings of the National Academy of Sciences 110, 2395–2400 (2013). dx.doi.org/10.1073/pnas.1213958110
Journal information: Small , Proceedings of the National Academy of Sciences
Provided by RIKEN