Pushing the boundaries of transcription: A new level of variation in messenger RNAs exposed
(Phys.org) —Like musicians in an orchestra who have the same musical score but start and finish playing at different intervals, cells with the same genes start and finish transcribing them at different points in the genome. For the first time, researchers at EMBL have described the striking diversity of messenger RNAs (mRNAs) that such start and end variation produces, even from the simple genome of yeast cells. Their findings, published today in Nature, shed new light on the importance of mRNA boundaries in determining the functional potential of genes.
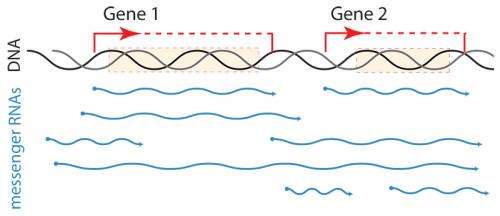
Hundreds of thousands of unique mRNA transcripts are generated from a genome of only about 8000 genes, even with the same genome sequence and environmental condition. "We knew that transcription could lead to a certain amount of diversity, but we were not expecting it to be so vast," explains Lars Steinmetz, who led the project. "Based on this diversity, we would expect that no yeast cell has the same set of messenger RNA molecules as its neighbour."
The traditional understanding of transcription was that mRNA boundaries were relatively fixed. While it has long been known that certain parts of mRNAs can be selectively 'spliced' out, this phenomenon is very rare in baker's yeast, meaning that the textbook one gene - one mRNA transcript relationship should hold. Recent studies have suggested that things aren't quite that simple, inspiring the EMBL scientists to create a new technique to capture both the start and end points of single mRNA molecules. They now discovered that each gene could be transcribed into dozens or even hundreds of unique mRNA molecules, each with different boundaries.
This suggests that not only transcript abundance, but also transcript boundaries should be considered when assessing gene function. Altering the boundaries of mRNA molecules can affect how long they stay intact, cause them to produce different proteins, or direct them or their protein products to different locations, which can have a profound biological impact. Diversifying mRNA transcript boundaries within a group of cells, therefore, could equip them to adapt to different external challenges.
The researchers expect that such an extent of boundary variation will also be found in more complex organisms, including humans, where some examples are already known to affect key biological functions. The technology to measure these variations across the entire genome as well as a catalogue of boundaries in a well-studied organism are a good starting point for further research. "Now that we are aware of how much diversity there is, we can start to figure out what factors control it," points out Vicent Pelechano, who performed the study with Wu Wei. Wei adds: "Our technique also exposed new mRNAs that other techniques could not distinguish. It will be exciting to investigate how these and general variation in transcript boundaries actually extend the functional capacity of a genome."
More information: Extensive transcriptional heterogeneity revealed by isoform profiling - Vicent Pelechano, Wu Wei, Lars M. Steinmetz – Advanced Online Publication in Nature on 24 April 2013 – DOI: 10.1038/nature12121
Journal information: Nature
Provided by European Molecular Biology Laboratory