How yeast responds to change: Development of new protocol provides key to measuring complete set of yeast protein levels
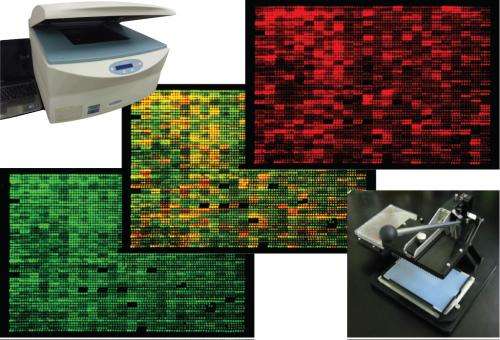
A procedure that allows accurate measurement of the levels of over 99% of the proteins generated by different strains of fission yeast could open the way for new laboratory applications of the species. Researchers should now be able to determine the direct impact of environmental change, life-cycle stage, and gene mutations, deletions and activity on this model organism.
The protocol, developed by Minoru Yoshida and Akihisa Matsuyama of RIKEN's Advanced Science Institute in Wako, together with colleagues from the University of Namur in Belgium, has successfully been manually implemented to study the effect of gene deletions. The researchers are confident, however, that it could easily be automated.
The advent of high-throughput genome sequencing has meant that complete genome sequences are readily available for many micro-organisms, allowing molecular biologists to deduce their protein sets—known as proteomes. This opens up the possibility of studying the proteome itself, and in particular how protein levels respond to changes in genes, age or the environment.
Fission yeast (Schizosaccharomyces pombe) has become a useful model for human genetic systems because it shares many similar genes and is easy to handle in the laboratory. The approximately 5000-gene fission yeast genome was first published in 2002, but several significant hurdles have prevented researchers determining accurate levels of all the proteins it generates. One obstacle was the yeast cell wall, which makes it difficult to extract proteins from the cell for analysis. In earlier work, Yoshida and colleagues successfully developed a means of bursting these cell walls to prepare mixtures of the proteins inside.
The new procedure allows comparison of the proteomes of different strains of yeast—typically a normal 'wild-type' strain with a genetic mutant or a strain raised in a specific environment. Specifically, it relies on tagging of open reading frames (ORFs)—the sections of chromosomes where proteins are encoded—with a small tag which is recognized by a fluorescently labeled antibody. The researchers introduced the tags to the strains of interest by means of a 'mass mating' from which only yeast progeny containing tagged ORFs were selected. The level of each protein could then be determined using a fluorescence scanner.
"We are now using the protocol to study the molecular mechanisms of ageing," says Matsuyama. "Proteins whose levels reproducibly alter after the yeast cells stop division can provide us with markers for age."
More information: Bauer, F., Matsuyama, A., Yoshida, M. & Hermand, D. Determining proteome-wide expression levels using reverse protein arrays in fission yeast. Nature Protocols 7, 1830–1835 (2012). www.nature.com/nprot/journal/v … /nprot.2012.114.html
Provided by RIKEN