Watching a protein as it functions
(Phys.org) —When it comes to understanding how proteins perform their amazing cellular feats, it is often the case that the more one knows the less one realizes they know. For decades, biochemists and biophysicists have worked to reveal the relationship between protein structural complexity and function, only to discover more complexity. One challenging aspect of protein behavior has been the speed with which they change shape and interact with their neighboring biomolecules. Until recently, researchers have relied on a somewhat static approach, using freeze-trapping to capture protein intermediates at various steps along a biochemical pathway. But exciting breakthroughs now allow us to watch proteins changing in real time.
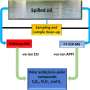
A research group has developed the necessary infrastructure at the BioCARS 14-ID-B beamline at the U.S. Department of Energy Office of Science's Advanced Photon Source to watch proteins function in real time on the picosecond time scale. Their work brings us many steps closer to knowing how proteins function, or malfunction when leading to disease.
A signaling protein usually responds to a messenger or trigger, such as heat or light, by changing its shape, which initiates a regulatory response in the cell. Signaling proteins are all-important to the proper functioning of biological systems, yet the rapid sequence of events, occurring in picoseconds, had, until now, meant that only an approximate idea of what was actually occurring could be obtained.
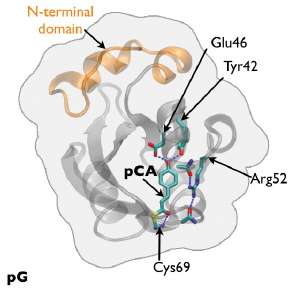
The researchers from the National Institutes of Health, the Nara Institute of Science and Technology (Japan), The University of Chicago, and the European Synchrotron Radiation Facility (France) worked to remedy that situation by developing a time-resolved, 150-ps Laue crystallography technique that can be utilized to watch structural changes in the photocycle of a protein first discovered in the photosynthetic bacterium Halorhodospira halophila. The protein, called photoactive yellow protein (PYP), is implicated in causing the bacterium to swim away from blue light, which could be genetically harmful, and toward green light, which it needs for photosynthesis.
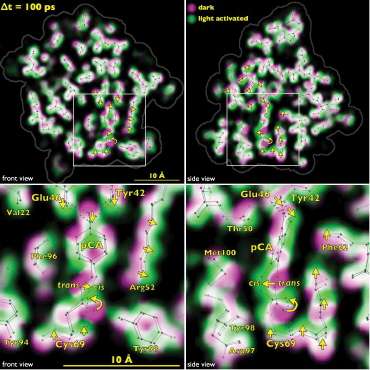
The team tracked the reversible photocycle of PYP over ten decades of time, from 100 ps to 1 sec. The signaling reaction is triggered by absorption of a photon of light by p-coumaric acid (pCA) chromophore, the non-protein entity that actually absorbs the photon, and which undergoes trans-to-cis isomerization. The bacterium responds to this light signaling within about half a second, indicating how rapidly the biochemical changes that facilitate the response must occur in the cell.
The team identified four major intermediates in the photoisomerization cycle. The first cis intermediate, lasting for only 600 ps, is in an unusual, highly contorted form not previously observed in crystallography experiments and is the precursor to intermediates that had been observed on the time scale of nanoseconds. Additional intermediate steps in the process of structural transition include breaking and making of hydrogen bonds, formation and relaxation of strain, and gated water penetration into the protein's interior. The researchers cross-validated their structural data, utilizing density functional theory calculations to confirm the chemical plausibility of the intermediates.
By tracking structurally the PYP photocycle with near-atomic resolution, the team provided a foundation for understanding the general process of signal transduction in proteins at nearly the lightning speed in which they are actually happening. By extending these techniques to enzymes, it would be possible to perform real-time observations of enzymatic action. Thanks to the team's groundbreaking work, the mechanistic and kinetic complexities with which proteins carry out their sophisticated functions are now much more clearly on view to those who have been eagerly waiting to see.
More information: Schotte, F. et al. Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography, Proc. Natl. Acad. Sci. USA 109(47), 19256 (November 20, 2012). DOI:10.1073/pnas.1210938109
Jung, Y. et al. Nat. Chem. 5, 212 (March 2013). www.nature.com/nchem/journal/v … full/nchem.1565.html
Provided by Argonne National Laboratory