Breakthrough could lead to drugs that better combat 'superbugs'
(Phys.org)—In the never-ending battle between antibiotic developers and the bacteria they fight, scientists at the U.S. Department of Energy's (DOE) Argonne National Laboratory have made a key breakthrough that could allow for the development of new drugs to more effectively combat antibiotic-resistant "superbugs."
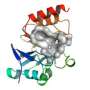
An Argonne team led by Youngchang Kim of the Structural Biology Center, in collaboration with researchers from the Midwest Center for Structural Genomics, the University of Texas-Pan American and Texas A&M University, recently determined the structure of NDM-1, a harmful enzyme able to overcome several antibiotics. The team used a combination of X-ray crystallography at Argonne's Advanced Photon Source (APS), biochemical assays, and computational modeling using resources at two Texas universities.
NDM-1, present in a number of pathogenic bacteria, including Klebsiella pneumonia and Escherichia coli, is able to defeat many of the world's most widely used antibiotics, including penicillin derivatives, cephalosporins, monobactams and carbapenems. The enzyme works by effectively binding to and breaking – in a process known as hydrolysis – a structure called a β-lactam ring, which is necessary for antibiotics to function.
The traditional view of enzymes, particularly those most essential for the bacteria to survive, is that they have a very specific substrate (or target – in this case the antibiotic) to bind and act on, and they interact with this target directly by forming a well-fitted lock-and-key-like complex. However NDM-1, and others like it, can act on a broad range of substrates.
"These kinds of enzymes can recognize many different targets," said Andrzej Joachimiak, head of Argonne's Structural Biology Center and the Midwest Center for Structural Genomics.
"The appearance of NDM-1 among pathogenic bacteria represents a major concern because the enzyme can inactivate so many of the antibiotics that we use to treat infections," said Charles Edmonds of the National Institutes of Health's National Institute of General Medical Sciences, which partially supported the study. "This work, by providing a detailed understanding of the structure of the enzyme and its mechanism of action, brings an invaluable tool to the design of new drugs to combat this significant threat to public health."
The act of NDM-1 binding to the antibiotic, however, does not represent the entire story, because for the enzyme to effectively overcome the antibiotic, it must also cleave the β-lactam ring. The researchers found that certain metals, including zinc, manganese and cadmium, can bind to the NDM-1 active site – and to the β-lactam ring. While zinc provides the most favorable environment for cleavage to occur, cadmium tends to inhibit the enzyme's ability to cleave the ring.
"The next step in the research is to look for inhibitors that we can create that would block the functioning of the enzyme," Joachimiak said. "If we can stop the enzyme from cutting the ring, the antibiotics stand a much better chance of staying effective."
The results of the research were recently published in The FASEB Journal in an article titled "NDM-1, the ultimate promiscuous enzyme: substrate recognition and catalytic mechanism."
Journal information: FASEB Journal
Provided by Argonne National Laboratory










.jpg)





