Understanding antibiotic resistance using crystallography and computation
.jpg)
(Phys.org)—Scientists at the University of Bristol, together with collaborators at the University of Aveiro, Portugal, have solved the structure of an enzyme that breaks down carbapenems, antibiotics 'of last resort' which, until recently, were kept in reserve for serious infections that failed to respond to other treatments.
Increasingly, bacteria such as E. coli are resisting the action of carbapenems by producing enzymes (carbapenemases) that break a specific chemical bond in the antibiotic, destroying its antimicrobial activity.
Carbapenemases are members of the group of enzymes called beta-lactamases that break down penicillins and related antibiotics, but it has not been clear why carbapenemases can destroy carbapenems while other beta-lactamases cannot.
Using molecular dynamics simulations, Professor Adrian Mulholland in the School of Chemistry and Dr Jim Spencer in the School of Cellular and Molecular Medicine, showed how a particular type of carbapenemase enzyme reorients bound antibiotic to promote its breakdown and render it ineffective.
Professor Mulholland said: "The class of antibiotics called carbapenems, drugs related to penicillin, are increasingly important in healthcare as treatments for bacterial infections. Until recently, carbapenems were 'antibiotics of last resort' but the growing problem of resistance to other drugs in organisms like E. coli (the leading cause of bloodstream infections in the U.K.) means that carbapenems are now becoming first-choice antibiotics for these infections. This is a worry because there are very few other treatment options for these organisms. Few new antibiotics effective against these pathogens are reaching the clinic.
"The recent appearance and spread of bacteria that resist carbapenems is a serious and growing problem: potentially, we could be left with no effective antibiotic treatments for these infections. The emergence of bacteria that resist carbapenems is therefore very worrying."
In a study published in the Journal of the American Chemical Society (JACS), the scientists combined laboratory experiments with computer simulations to investigate how one particular type of carbapenemase recognises and breaks down antibiotics.
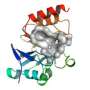
Using X-ray crystallography, they obtained two 'snapshots' of the carbapenemase in the act of breaking down a carbapenem antibiotic. This static structural information was used as a starting point for simulations that modelled the motions of the enzyme and the bound antibiotic.
The simulations showed how the carbapenemase reorients the drug to promote its breakdown. In beta-lactamases that cannot break down carbapenems, this rearrangement cannot happen, and so the enzyme cannot break down the antibiotic. Knowing this should help in designing new drugs that can resist being broken down.
Dr Spencer said: "Combining laboratory and computational techniques in this way gave us a full picture of the origins of antibiotic resistance. Our crystallographic results provided structures which were the essential starting point for the simulations and the simulations were key to understanding the dynamic behaviour of the enzyme-bound drug.
"Identifying the molecular interactions that make an enzyme able to break down the drug, as we have done here, is an important first step towards modifying the drug to overcome bacterial antibiotic resistance."
More information: 'The Basis for Carbapenem Hydrolysis by Class A β-Lactamases: A Combined Investigation using Crystallography and Simulations' by Fátima Fonseca , Ewa I Chudyk , Marc Willem van der Kamp , António Correia , Adrian J. Mulholland , and James Spencer in JACS (Journal of the American Chemical Society) pubs.acs.org/doi/abs/10.1021/ja304460j
Journal information: Journal of the American Chemical Society
Provided by University of Bristol