Penn researcher part of $1.5 million grant to reduce gene sequencing costs
A collaboration between researchers at Columbia University and Marija Drndić of the University of Pennsylvania has been awarded a three-year, $1.5 million grant for a project aimed at reducing the cost of genome sequencing. The grant was made by the National Human Genome Research Institute, part of the National Institutes of Health.
Drndić is an associate professor in the Department of Physics and Astronomy in Penn's School of Arts and Sciences.
During the past decade, DNA sequencing costs have fallen dramatically, fueled by tools, technologies and process improvements developed by genomics researchers.
Price, however, is still one hurdle in the widespread use of genomics in research and clinical care. Speed and accuracy are among other factors. The grants, made through the NHGRI's Advanced DNA Sequencing Technology Program and totaling $19 million between six groups, will attempt to address all of these challenges.
"We can now access data we could not dream of getting in 2004 when we started this program; tens of thousands of human genome sequences have been generated," said NHGRI director Eric D. Green. "And yet, the information we would truly like to get for understanding disease and, eventually, for treating patients, requires much better quality sequence data. That is the direction we would like to go with these grants."
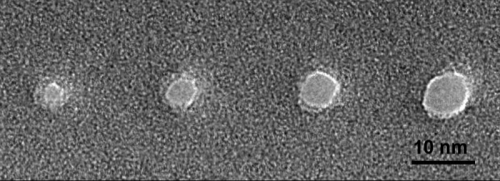
Drndić's approach to gene sequencing involves a process known as nanopore translocation, in which the DNA is threaded through a tiny hole. The elements, or bases, of a DNA sequence block different amounts of the aperture as they pass through, enabling the nanopore to emit a different signal for each base.
But the signals from nanopores are very weak, so it is critically important to measure as cleanly as possible. Collaborating with a research team led by Columbia's Ken Shepard, Drndić miniaturized the measurement by designing a custom-integrated circuit using commercial semiconductor technology, building the nanopore measurement around the new amplifier chip. Their research was published in the journal Nature Methods.
"While most groups were trying to slow down DNA, our approach was to build faster electronics," Drndić said. "We combined the most sensitive electronics with the most sensitive solid-state nanopores."
The NHGRI grant will help advance the study of this technique and provide for new equipment, which in turn will help improve the speed and accuracy of the group's sequence measurements.
Drndić is also the recipient of a 2011 NHGRI/NIH grant for related work on genome sequencing using graphene nanopore translocation.
Journal information: Nature Methods
Provided by University of Pennsylvania

















