Gene regulatory proteins may be more flexible in their DNA binding preferences than previously expected
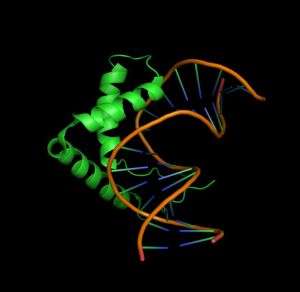
Transcription factor proteins essentially act as genetic on/off switches, binding specific stretches of DNA that enable them to stimulate or repress the activity of neighboring genes. However, it remains a challenge to authoritatively define the DNA sequences individual factors prefer.
Work from Prasanna Kolatkar’s team at the A*STAR Genome Institute of Singapore now offers valuable insights into this process. The human genome encodes 20 members of the Sox transcription factor family, which play a key role in many developmental processes, and Kolatkar and colleagues are exploring how these proteins recognize their respective targets.
Previous experiments have defined a core TTGT binding sequence for Sox proteins, but other determinants are clearly involved. The researchers began by comparing the structures of the complexes formed by the DNA-binding ‘high mobility group’ (HMG) domains of either Sox4 or Sox17 with a specific target DNA sequence. Both proteins formed highly similar complexes in these experiments, although there were subtle differences in the Sox4 HMG-DNA interaction that prompted closer investigation.
In follow-up experiments, the researchers assessed the binding efficiency of the Sox4 HMG against a broad variety of DNA sequences. To their surprise, they learned that although this domain preferentially binds to a specific ‘primary motif’, it can also interact strongly with various ‘secondary motifs’ (see image). All of these motifs contain the TTGT core, but with variations in the flanking sequences. Significantly, Kolatkar’s team noted a ‘positional interdependence’ effect within these flanking sequences, such that Sox4 HMG required the presence of specific nucleotide pairs to bind effectively — for example, favoring CT or AA over other combinations.
The researchers determined that the subtle differences they had observed in the Sox4 binding complex actually represented the amino acids that enabled Sox4 to sense and discriminate between its primary and secondary binding sites. “The same protein can recognize two separate DNA motifs for binding with different affinities using slight changes at the interaction surface,” says Kolatkar.
These findings suggest that transcription factors in general may have greater flexibility with regard to their site preferences than was previously recognized, and Kolatkar’s team is now delving more deeply into the interaction behavior of the rest of the Sox family. If this model is confirmed, it could represent an important system for control of gene expression; for example, certain genes containing ‘secondary’ regulatory motifs may only get switched on when transcription factor levels are especially high. “This could help us to rationalize the mechanisms underlying genomic regulatory network data,” says Kolatkar.
More information: Jauch, R., Ng, C. K., Narasimhan, K. & Kolatkar, P. R. Crystal structure of the Sox4 HMG/DNA complex suggests a mechanism for the positional interdependence in DNA recognition. Biochemical Journal 443, 39–47 (2012). dx.doi.org/10.1042/BJ20111768


















