Combining crystallography and visible spectroscopy to understand enzymes
Structure and function are intimately linked, but do not necessarily predict the other. For example, x-ray crystallography provides 3D atomic structural information about biological macromolecules but does not define important details about metal ions.
However, the oxidation state of metal ions at an enzyme’s active site has a critical effect on enzyme behavior. Thus, an enzyme’s catalytic function derives from the electronic structure of those atoms influencing or directly participating in the reaction, information not revealed by the scattering methods used in x-ray crystallography.
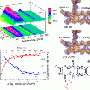
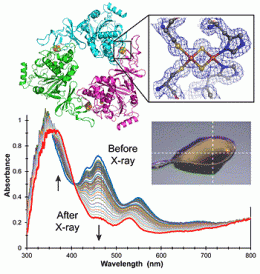
A new technology has been developed that simultaneously carries out crystallography and UV-visible and Raman spectroscopy to determine the atomic structure of the entire protein, and electronic and vibrational structures of the metal ions or cofactors within. The combined instrumentation has been used to study the process of demethylation of an organic substrate molecule by an enzyme whose active site includes an iron-sulfur cluster.
The researchers used spectroscopy to follow the change in the oxidation state of the cluster during the crystallography data collection, and to formulate a mechanism for the overall enzyme reaction mechanism. The results provide insight into a new demethylation reaction, an important class of phenomena that control cellular behavior.
The technology was developed by scientists at the Protein Crystallography Research Resource at the National Synchrotron Light Source at Brookhaven National Laboratory. The new study was led by Allen M. Orville of Brookhaven and Pinghua Liu and Karen N. Allen of Boston University, and is published in the Journal of the American Chemical Society.
Journal information: Journal of the American Chemical Society
Provided by Brookhaven National Laboratory