Another piece of the ion pump puzzle
From bacteria to humans, all cells use molecules of adenosine triphosphate (ATP) as fuel to power a broad range of biochemical reactions. For example, massive multi-subunit enzymes known as V-ATPases convert ATP molecules into energy that helps drive the transport of ions across cellular membranes.
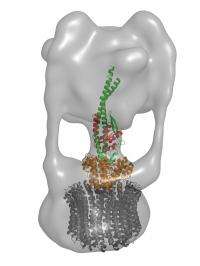
V-ATPases consist of a so-called ‘V complex’, which transfers energy derived from ATP hydrolysis into rotational motion, thereby promoting ion transport through to the membrane-bound V0 complex. These two complexes are joined by three ‘stalks’, including a central stalk composed of subunits named D and F, although this segment of the protein is poorly characterized. “The structure of this central axis of V-ATPase has not been obtained,” says Takeshi Murata of the RIKEN Systems and Structural Biology Center in Yokohama, “and we believe such structural studies are very important to understand this protein’s precise mechanism.”
Murata and colleagues recently succeeded in obtaining high-resolution structural information about the DF complex of V-ATPase obtained from the bacteria Enterococcus hirae1. By comparing this structural information against an equivalent segment from F-ATPase, which synthesizes rather than hydrolyzes ATP, the researchers were able to identify functional domains that may be specifically required by V-ATPases.
They determined that the E. hirae D subunit is composed of a pair of long helical structures coiled around each other, with a short hairpin-shaped loop at one end. According to Murata, the discovery of this latter structure was unexpected. “This short beta-hairpin region is a unique structure, although the rest of the D structure is very similar to that of other rotary complexes such as F-ATPase and flagellar motors,” he says. This segment does not appear to be essential for V-ATPase assembly, but ATP processing efficiency was reduced when the researchers deleted this hairpin from the subunits.
In contrast, the E. hirae F subunit assumed a more compact structure, relatively similar to its A- and F-ATPase counterparts; the researchers determined that it specifically associates with the middle portion of the D subunit’s coiled helical segment, an interaction that depends heavily on a particular helix within the F subunit.
Although untangling this structure represents a major step forward, this complex must also be understood as part of a far larger entity (Fig. 1). Murata and colleagues have already begun tackling this. “We recently succeeded at solving the structure of V1-ATPase with a resolution of 2.1 Angstroms,” says Murata, “and we are now preparing this manuscript for publication.”
More information: Saijo, S.,et al. Crystal structure of the central axis DF complex of the prokaryotic V-ATPase. Proceedings of the National Academy of Sciences USA 108, 19955–19960 (2011).
Provided by RIKEN





















