New resource opens the door for enzyme research
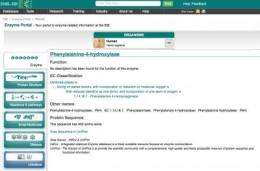
The European Molecular Biology Laboratory's European Bioinformatics Institute has launched the Enzyme Portal, a freely available resource for people who are interested in the biology of enzymes and proteins with enzymatic activity.
Enzymes catalyse the myriad reactions that take place in living organisms, allowing chemical changes to occur that would otherwise need conditions that are incompatible with life. Until now, information about enzymes was scattered throughout many different resources. This meant that you had to know exactly what you were looking for, which could be a real barrier to discovery.
The Enzyme Portal mines and displays data about proteins with enzymatic activity from public repositories via a single search, and includes biochemical reactions, biological pathways, small molecule chemistry, disease information, 3D protein structures and relevant scientific literature. It summarises information in the UniProt knowledge base; the Protein Data Bank in Europe; Rhea, a database of enzyme-catalysed reactions; Reactome, a database of biochemical pathways; IntEnz, a resource with enzyme nomenclature information; ChEBI and ChEMBL, which contain information about small-molecule chemistry and bioactivity; and CoFactor and MACIE for highly detailed, curated information about cofactors and reaction mechanisms.
The Enzyme Portal covers a large number of species, including the key organisms used in biological research, and makes it simple to compare the characteristics of equivalent enzyme activities in different organisms.
"The Enzyme Portal will serve researchers interested in the native metabolism of organisms as well as those working in drug discovery or chemical biology," explained Dr Christoph Steinbeck, Head of Cheminformatics and Metabolism at EMBL-EBI. "The resource seamlessly bridges the various enzyme-related aspects of these areas, all the way from the small molecules – which may occur naturally in the organism of study or be introduced – to the 3D structure of the enzymes affected and the genomic information coding for those."
The design of the Enzyme Portal was based entirely on user demand and feedback. "We didn't come in with preconceived ideas of what it should be; it was designed by users, for users," said Cheminformatics Coordinator Paula de Matos. "We had enough data, but it was distributed over 10 different resources. Now, we have a central place where they can access and explore all of it – including information about disease."
"The Enzyme Portal takes the researcher's perspective," added User Experience Analyst Jenny Cham. "We leveraged user-testing feedback to create it, making it the first EMBL-EBI resource to have a fully user-centred design from scratch. This approach was not only cost-effective; it made decision making and communication much easier – particularly in terms of design and technology choices."
Provided by European Molecular Biology Laboratory