Chemists mimic nature to design better medical tests

Over their 3.8 billion years of evolution, living organisms have developed countless strategies for monitoring their surroundings. Chemists at UC Santa Barbara and University of Rome Tor Vergata have adapted some of these strategies to improve the performance of DNA detectors. Their findings may aid efforts to build better medical diagnostics, such as improved HIV or cancer tests.
Their research is described in an article published this week in the Journal of the American Chemical Society.
Nature often serves as a source of inspiration for the development of new technologies. In the field of medical diagnostics, for example, scientists have long taken advantage of the high affinity and specificity of biomolecules such as antibodies and DNA to detect molecular markers in the blood. These molecular markers allow them to monitor health status and to guide treatments for diseases, including HIV, cancer, and diabetes.
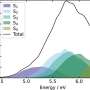
Kevin W. Plaxco, a professor of chemistry at UCSB, whose group carried out the research, notes that despite their great attributes, a main limitation of such biosensors is their precision, which is confined to a fixed, well-defined "dynamic range" of target concentrations. Specifically, the useful dynamic range of typical biomolecule binding events spans an 81-fold range of target concentrations
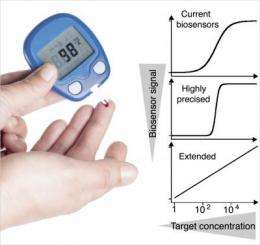
"This fixed dynamic range complicates –– or even precludes –– the use of biosensors in many applications," said Plaxco. "To monitor HIV progression and provide the appropriate medication, for example, physicians need to measure the levels of viruses over five orders of magnitude. Likewise, the two orders-of-magnitude range displayed by most biosensors is too broad to precisely monitor the concentrations of the highly toxic drugs used to treat many cancers. Our goal was, therefore, to create sensors with extended (for applications needing a broad dynamic range) or narrowed (for applications needing high measurement precision) dynamic ranges at will."
The key breakthrough underlying their new approach came from the simple observation of nature. "All living organisms monitor their environments in an optimized way by using sensing molecules that respond to either wide or narrow change in target concentrations," said Alexis Vallée-Bélisle, a postdoctoral fellow and the first author of the study. "Nature does so by combining in a very elegant way multiple receptors, each displaying a different affinity for their common target".
Inspired by the optimized behaviors of these natural sensors, the UCSB research group teamed up with Francesco Ricci, professor at the University of Rome Tor Vergata to do their own mixing and matching of biomolecules to manipulate biosensors' dynamic ranges. To validate their approach, they used a widely employed DNA-based biosensor used for detecting mutations in DNA called a "molecular beacon."
By combining sets of molecular beacons all binding the same target molecule but with differing affinities, the international team was able to create sensors with rationally "tuned" dynamic ranges. In one case, they developed a sensor that monitors DNA concentrations over a six orders of magnitude range. In another example, they developed an ultrasensitive sensor that precisely detects small changes in target concentration over only a five-fold dynamic range. Finally, they also built sensors characterized by complex, "custom-made" dynamic ranges in which the sensor is insensitive within a window of desired concentrations (e.g., the clinically "normal" concentration range of a drug) and very sensitive above or below this "appropriate" concentration range. The researchers believe that these strategies can be in principle applied to a wide range of biosensors, which may significantly impact efforts to build better point-of-care biosensors for the detection of disease biomarkers.
Provided by University of California - Santa Barbara