Study provides new insights into an ancient mechanism of mammalian evolution
A team of geneticists and computational biologists in the UK today reveal how an ancient mechanism is involved in gene control and continues to drive genome evolution. The new study is published in the journal Cell.
To function properly, mammalian tissues require the protein CTCF, which has several key activities including the regulation of genes and interaction with proteins in the cell's nucleus to alter gene activity. CTCF acts by binding to DNA and plays a role in diseases such as HIV infection and cancer. However, very little is known about the origin of the DNA sequences that are bound by CTCF.
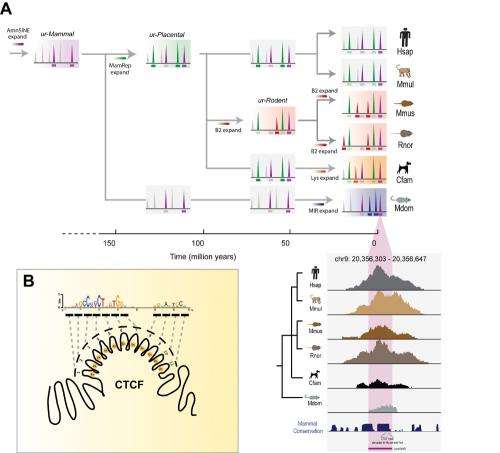
In this study, the researchers used samples from six mammals (human, macaque, mouse, rat, dog, and short-tailed opossum) to pinpoint where CTCF binds to each genome. They discovered around 5000 sites that are present in most cell types and tissues, and that have not changed over hundreds of millions of years of mammalian evolution. Because these CTCF binding sites are conserved throughout evolution, the researchers believe that many might play an important role in gene regulation.
The team found an even larger number of locations where CTCF binds DNA in only one lineage or a single species. These additional sites represent a signature of important evolutionary changes since our last common ancestor – legacies, in some cases, of the evolutionary path to humans. These newer CTCF sites are embedded inside virus-like stretches of DNA called 'retro-transposons'. Retro-transposons use a copy-paste mechanism to spread copies of themselves throughout the genome.
"We developed a new, integrated model of CTCF evolution, which explains the origin of these 5000 highly conserved CTCF binding events in mammals," said Paul Flicek of the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) and the Wellcome Trust Sanger Institute. "Taken together, our findings provide fascinating insight into an ancient mechanism of evolution that is still actively changing our genome."
"CTCF is a key regulator involved in chromatin and gene expression remodelling, both of which are perturbed in the development of cancer. The gene expression and chromatin changes in cancer have also recently been relied on to predict the outcome of specific cancer treatments, which is why it is so important to have a detailed understanding of how particular parts of the genome are resistant or plastic to changes," said Duncan Odom of Cancer Research UK and the Wellcome Trust Sanger Institute.
The retro-transposon's copy-and-paste behaviour has long been considered totally self-serving. However, the study showed that when a retro-transposon containing a CTCF-binding sequence spreads around a mammal's genome, it can deposit functional CTCF binding sites in novel locations, altering the activity of distant genes.
"We looked at six mammalian species representing primates, marsupials, rodents and carnivores, and discovered a simple mechanism that they all use to remodel their DNA," explained Petra Schwalie of EMBL-EBI. "We also found that our distant ancestors also experienced the same complicated relationship between CTCF and retro-transposons."
Using molecular palaeontology techniques, the researchers were able to identify fossil traces of older retro-transposon expansions in the DNA around the shared CTCF binding locations, and showed that this process has been active for hundreds of millions of years.
More information: Waves of repeat-driven CTCF binding expansions have shaped mammalian genomes. Schmidt D., Schwalie P., Wilson M.D., et al. Cell 2012, DOI: 10.1016/j.cell.2011.11.058 dx.doi.org/10.1016%2Fj.cell.2011.11.058
Journal information: Cell
Provided by European Molecular Biology Laboratory















