High-resolution mapping of the 3D organization of chromosomes
In collaboration with researchers from the Weizmann Institute in Israel, a team from the Institut de Génétique Humaine (CNRS, France) has, for the first time, revealed the detailed three-dimensional architecture of chromosomes: Giacomo Cavalli and his colleagues have achieved high-resolution mapping of the different contacts that exist within and between chromosomes. They performed this feat using a new very high-throughput technique improved by the Montpellier team. This major research work should shed new light on the impact of 3D chromosome organization on genome expression and on the onset of diseases such as cancer. It is published in the online version of the journal Cell of 19 January 2012.
Researchers had suspected for years: individuals are more than the sum of their genes and therefore the architecture of their chromosomes could also be a key factor in the proper functioning of their cells. However, until now, a real understanding of the principles governing the folding of the genome in the nucleus was beyond the scope of existing approaches. Consequently, for the last four years, twenty or so research teams in various parts of the globe have been developing methods to study genome folding.
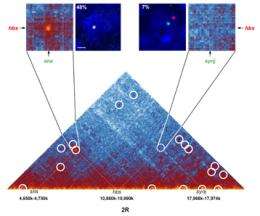
To describe interactions between genomic regions, the CNRS team used a method known as “chromosome conformation capture” (Hi-C), derived from a technique developed in 2009 by an American group. Like the American version, the Montpellier method involves three major steps: fixing intra- and inter-chromosomal interactions with formaldehyde; binding the ends of the two regions in contact with the DNA ligase protein, of bacterial origin; and determining the sequence of these two contacting regions. Yet Cavalli and his colleagues brought significant improvements to the technique, making it possible to study much smaller fragments of DNA (several hundred base pairs, compared to fragments of 5-10 kilobases previously), and thus allowing these interactions to be mapped at high resolution.
Using this technique, the Montpellier researchers and their Israeli colleagues analyzed the genome of Drosophila (also known as fruit fly), an insect whose genome is fifteen times smaller than our own. The team was able to identify no fewer than 118 million contact zones! Analysis of these zones enabled the scientists to produce very detailed interaction maps. These show that the chromosomes are organized into domains containing one or more genes. Separated by chromatic boundary regions known as "insulators", these domains correspond either to actively expressed zones, or to inactive regions.
The mathematical expertise of Amos Tanay's Israeli team helped both groups develop a computer model capable of predicting, with quite good accuracy, the real folding of chromosomes. This very powerful tool has made it possible to elucidate the fundamental principles underlying the 3D organization of the genome.
In particular, it appears that the inactive domains tend to form interactions with other inactive domains, often nearby on the same chromosome, whereas active domains “contact” other active domains, either close or distant, in some cases on other chromosomes.
This crucial work should provide better understanding of the differences in chromosomal folding between different types of cells (liver cells, muscle cells, nerve cells, etc.), and the importance of such folding in the onset of cancer.
More information: Three-dimensional folding and functional organization principles of the Drosophila genome. Giacomo Cavalli et al. Cell. 19 January 2012 (on-line version)
Journal information: Cell
Provided by CNRS















