Similarities cause protein misfolding
A large number of illnesses stem from misfolded proteins, molecules composed of amino acids. Researchers at the University of Zurich have now studied protein misfolding using a special spectroscopic technique. Misfolding, as they report in Nature, is more frequent if the sequence of the amino acids in the neighboring protein domains is very similar.
A large number of illnesses stem from misfolded proteins, molecules composed of amino acids. Researchers at the University of Zurich have now studied protein misfolding using a special spectroscopic technique. Misfolding, as they report in Nature, is more frequent if the sequence of the amino acids in the neighboring protein domains is very similar.
Proteins are the main molecular machines in our bodies. They perform a wide range of functions, from digesting and processing nutrients, converting energy and aiding cell structure to transmitting signals in cells and the whole body. In order to perform these highly specific functions, proteins have to adopt a well-defined, three-dimensional structure. Remarkably, in most cases they find this structure unaided once they have been formed out of their individual building blocks, amino acids, as a long chain molecule in the cell.
However, the process of protein folding can also go wrong, which means the proteins affected are no longer able to perform their function. In some cases, this can even have much more serious consequences if these misfolded proteins clump and trigger neurodegenerative diseases such as Alzheimer's or Parkinson's disease.
In the course of evolution, a crucial factor in the development of proteins has thus been to avoid such "misfolding processes". However, this is no easy task since the same molecular interactions that stabilize the correct structure of the individual proteins can also bring about interactions between protein molecules, causing them to misfold.
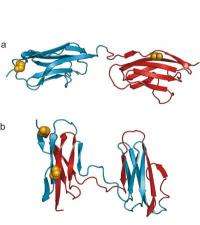
Using a special spectroscopic method called single-molecule fluorescence, researchers from the Universities of Zurich and Cambridge have now studied the circumstances under which misfolding occurs. The team headed by Prof. Benjamin Schuler from the University of Zurich studied sections, or "domains", of the largest protein in our bodies, titin, which helps the stability and elasticity of the muscle fibers. It is assumed that individual titin domains can unfold while the muscle is heavily exerted to avoid damaging the muscle tissue. When the muscle relaxes again, however, there is a danger that these unfolded domains might fold incorrectly. There is also a similar risk for other multidomain proteins.
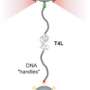
For their study, the researchers attached small dye molecules as probes in the protein. "Using our laser-spectroscopic method we were able to determine distances on a molecular scale, i.e. down to a few millionths of a millimeter, through the energy transfer between the probes," explains Prof. Schuler. This enabled the structures of correctly and misfolded proteins to be distinguished and thus the proportion of misfolding determined.
"The study of different titin domains in our experiments revealed that the probability of misfolding increases if neighboring domains are very similar in the sequence of their amino acids," says Prof. Schuler. This is apparently the reason why neighboring domains in proteins have a limited degree of similarity. "This seems to be a key evolutionary strategy to avoid protein misfolding and thus guarantee their maximum functionality," says Schuler.
Provided by University of Zurich