How to separate a sheep from its flock
(PhysOrg.com) -- When the signaling proteins known as kinases stop working, the results can be dramatic. Glitches in the enzymes can trigger diabetes, impair immune function, or drive the spread of cancers, and scientists are finding more and more examples of kinase mutations that drive disease. It’s not surprising that the list of guilty kinases is long, because the family of enzymes is large – in healthy cells, more than 500 different kinases help keep all manner of essential processes running smoothly.
Howard Hughes Medical Institute investigator Kevan Shokat has dedicated his lab to devising new ways of studying individual kinases within this closely related family. His methods have been adopted by other labs and helped clarify the role of more than 70 human kinases.
Because they are enzymes -- proteins that catalyze reactions in cells – kinases are generally considered good drug targets. In theory, drug developers can design molecules that exploit an enzyme’s unique structure or mode of action to block it without harming other players within the cell. But the sheer numbers of kinases inside a cell presents a challenge when researchers want to single one out for study.
Typically, to discover the role of a particular player within a chain of proteins, scientists are likely to try disabling it. That strategy has been crucial, for example, in understanding how some oncogenes hijack the signaling that controls normal cell division. But what if the protein in question is indistinguishable from dozens, even hundreds, of others? You can’t learn how enzyme A works if blocking its action also disables enzymes B through Z. Not only is it futile, but it’s a sure way to kill a cell. For that reason, kinases, which share strong structural similarities to one another, can leave the standard tools of inhibitor designers a bit flummoxed.
Kinases account for some two percent of the human genome. They transfer energy-bearing phosphates from ATP to proteins, advancing crucial molecule-to-molecule relay races that drive everything from cell division and development to learning and memory.
When the relay runs smoothly, all is well. But a mutation in one protein along the chain can disrupt the signals and wreak havoc.
In 1997, Shokat saw a way through the conundrum of kinase research. He developed methods to zero in on individual kinases for study, and to block them if necessary to treat disease.
Shokat, whose lab is at the University of California San Francisco, figured out that the best way to find the differences between kinases was to focus on what they have in common. He knew that all kinase molecules contain a sculpted pocket that accepts an ATP molecule. ATP docks at this site and delivers phosphate, a chemical that acts as an on/off switch for many enzymes.
Shokat’s idea is called chemical genetics: he decided to mutate a particular kinase so that its binding pocket no longer made a perfect fit for ATP. He reshaped the pocket to accept a drug-like molecule instead. Depending on the type of drug used, Shokat could block normal ATP binding and either disable the kinase or simply tag it so it could be distinguished from all other kinases.
This chemical change to the pocket made a big difference. It enabled Shokat to single out one kinase for study, distinguishing it from all its nearly identical cousins. And it could be done without interfering with ATP’s job of delivering phosphate fuel to all the other kinases in the cell.
Other methods developed in Shokat’s lab let researchers tweak a kinase so that it becomes that only kinase in a cell that no longer cooperates only with standard ATP, but works also with a chemically labeled ATP supplied by the researchers. Phosphates taken from these labeled ATPs are easily traceable, so the scientists can determine exactly where the kinase they are studying puts them – in other words, which other proteins within a cell are targets of the kinase.
By tagging one kind of kinase at a time, researchers can now figure out the role of kinases in cell signaling cascades. The strategy allows scientists to learn, for example, whether a specific kinase among half a dozen enables an oncogene to take over cell division. Because the technique provides a window on normal protein interactions in cells, it can pinpoint where researchers might interrupt the flow of protein-to-protein signaling to treat disease. A kinase inhibitor initially developed by his lab is now in clinical trials to treat cancer, and shows promise in easing other diseases as well.
After 17 years these kinds of studies, Shokat remains stunned by the complexity of the actions kinases drive. “It really blows me away that hundreds of kinases act on a millisecond timescale in order for a cell to decide to grow or not grow, or for a cancer cell to grow out of control,” he says.
Most recently, in work published online March 27, 2011, in the journal Nature, Shokat has collaborated with crystallographer David Barford of London’s Institute of Cancer Research to investigate a protein called KSR (kinase suppressor of Ras).
For 15 years, scientists have known that KSR plays a role in a signaling pathway that drives the spread of about one-third of all cancers. But they had not been able to determine how it did so.
About 90 percent of all pancreatic cancers and about half of thyroid and colorectal cancers are caused by an oncogene that locks an enzyme called RAS in the ‘on’ position. Ras transmits its growth-spurring signal via two kinases, Raf and MEK. Clinical trials are currently underway evaluating drugs that aim to treat cancer by blocking these kinases.
Despite intense study of the Ras pathway by many labs, much about it remains unknown. Shokat knew that more information might help drug developers design more effective drugs or drugs with fewer side effects. So he integrated his chemical genetics approach with structural information gleaned by Barford’s lab to produce a much clearer picture of how KSR helps the other two kinases, Raf and MEK, work together to trigger cell growth. Work from other labs had clarified that KSR acts as a structural scaffold that brings members of the Ras pathway together so that they can interact, and although KSR looked like a kinase, some researchers suspected its scaffolding function might be KSR’s sole role.
As a scaffold, KSR might be difficult to disrupt, but if it also acts as a kinase, it is a more promising drug target. So Barford and Shokat set out to determine whether KSR could transfer a phosphate signal to members of the Ras pathway during their association, and to show how a signal from KSR would affect other kinases’ behavior.
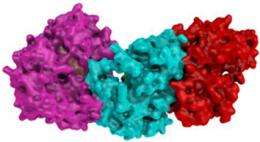
Using x-ray crystallography, Barford’s group determined the structure of KSR when it is assembled in a complex with the kinase MEK. From the molecular picture they produced, it was clear that KSR was well positioned to link a phosphate onto MEK.
Using Shokat’s chemical genetic technique, the team went on to demonstrate that KSR can indeed act as a kinase, transferring a phosphate molecule to MEK. Their experiments indicated that Raf spurs KSR on by altering KSR’s shape, and that this shape shift helps transmit the oncogene’s excessive growth signal to MEK.
Since 2009 alone, the Shokat lab’s research findings have been published in more than three dozen papers. But Shokat says this latest finding has been particularly energizing.
“To me, this is the most exciting discovery we’ve published so far,” he says. “RAS is a pernicious oncogene. It is the most frequently activated gene in cancer, but it’s undruggable. Now we have a much clearer understanding of how it works, and the prospect of making a drug against 35 percent of all cancers.”
He’s been at the kinase game long enough to know that new insights about signaling pathways don’t lead directly to the jackpot of a killer cancer drug. But as he puts it, “I hope there are only a few twists and turns left along the way.”