Protein-devouring enzyme complex uses two different mechanisms to determine which targets to destroy
The proteasome is the garbage-disposal system of the cell, enzymatically clearing away unwanted proteins. Since this requires the recognition of individual targets within the crowded cellular environment, it is critically important that molecules ‘marked for death’ are appropriately flagged.
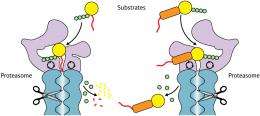
This signal, known as the degron, is composed of two components: an unstructured ‘initiation region’ within the target protein and a proteasome recognition tag. This tag typically consists of a chain of ubiquitin molecules, but some proteins get steered to the proteasome with the help of ubiquitin-binding ‘adaptor’ proteins. “These two pathways work in parallel with and independently from each other, and converge at the initiation step,” explains Tomonao Inobe of the RIKEN Brain Science Institute in Wako, Japan.
By analyzing the efficiency with which different synthetic protein constructs get degraded by the proteasome, Inobe and colleagues in Andreas Matouschek’s laboratory at Northwestern University in Illinois, USA, have uncovered important structural details of the recognition mechanisms used by the proteasome to manage these distinct pathways.
The team’s initial experiments showed that the minimum length for the initiation region is shorter in proteins tagged with ubiquitin alone (Ub4) than those tagged with an adaptor-derived ubiquitin-like (UbL) domain. Similarly, they found that Ub4–mediated degradation was most efficient when these sites were close together, and was impaired by the insertion of rigid ‘spacer’ protein segments between the two degron components. With UbL-tagged constructs, however, degradation was maximized when these components were moderately separated.
Based on their data, the researchers concluded that these physical constraints arise because Ub4- and UbL-tagged proteins bind to completely different sites on the proteasome; ubiquitin binds very near to the digestion machinery, requiring the initiation region to be close by (Fig. 1), while the UbL-binding site is considerably farther away, and thus accommodates greater separation. Inobe compares this to how an electrical plug must match its outlet. “The proteasome can recognize different plugs,” he says, “but each one has to have the correct specific arrangement of prongs.”
Inobe hopes to better characterize the functional role of this distance restriction in the future, but suggests that this mechanism may enable this protein complex to achieve both direct destruction of individual proteins and the targeted degradation of specific molecules nestled within larger complexes. “The spacing rules fit well with the way these tags are used physiologically and help explain how substrates are selected for degradation or manage to escape the process,” says Inobe.
More information: Inobe, T., et al. Defining the geometry of the two-component proteasome degron. Nature Chemical Biology 7, 161–167 (2011). www.nature.com/nchembio/journa … bs/nchembio.521.html
Provided by RIKEN
















