Key difference in how TB bacteria degrade doomed proteins

(PhysOrg.com) -- Scientists at the U.S. Department of Energy's (DOE) Brookhaven National Laboratory and Stony Brook University have discovered a key difference in the way human cells and Mycobacterium tuberculosis bacteria, which cause TB, deliver unwanted proteins — marked with a "kiss of death" sequence — to their respective cellular recycling factories. This critical difference, described in a paper published online October 17, 2010, in the journal Nature Structural and Molecular Biology, may help scientists design drugs to disable the bacterial system while leaving normal human protein recycling centers intact.
"With tuberculosis infecting a third of the world's population, primarily in developing countries, there is great need for new, effective TB treatments," said study co-author Huilin Li, a Brookhaven biophysicist and associate professor at SBU. "Our research seeks to understand the protein-recycling mechanism of TB bacteria, because it is one of the microbe's keys to survival in human cells. Targeting this system with small-molecule-based drugs could inhibit the bacteria and effectively treat TB."
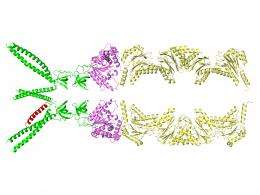
The catch is that human cells have a similar protein-recycling system, essential for their survival, which could also be destroyed by inhibitory drugs. "It's important to find differences between the species so we can target features unique to the bacterial system," Li said.
Li has previously looked at differences in the cellular structure known as a proteasome that chops up the unwanted proteins [see links below]. The current study examined the way proteins destined for degradation are recognized by the bacterial proteasome before entering that structure.
Using beams of high-intensity x-rays at the Lab's ["www.nsls.bnl.gov/">National Synchrotron Light Source (NSLS), the scientists determined atomic-level structures of the portion of the bacterial proteasome that identifies the unwanted protein's "kiss of death" marker sequence — as well as structures of the marker sequence as it binds with the proteasome.
Based on the structures, the scientists describe a detailed mechanism by which coiled, tentacle-like arms protruding from the proteasome identify the death sentence label, causing a series of protein-folding maneuvers that pull the doomed protein into the degradation chamber.
Importantly, this interaction between the bacterial proteasome and the marker sequence is unique to bacteria. Human cells use a different marker protein and a completely different mechanism for drawing doomed proteins into the proteasome. Thus the details of proteasome-substrate interaction revealed by the current study may provide highly specific targets for the development of new anti-tuberculosis therapies.
Provided by Brookhaven National Laboratory















