Scientists post lower speed limit for cell-signaling protein assembly
The apparently random self-assembly of molecular threads into the proteins that make the body work is far less frantic than previously thought, Michigan State University scientists say. That discovery could be a key to help unlock the nature of some diseases.
How proteins spontaneously "fold" from wiggling chains of amino acids into a wide variety of functional - or malfunctioning - three-dimensional molecules is one of the biggest mysteries in biochemistry.
"People thought they understood how protein diffusion worked, but now our data suggests they're wrong by a factor of 1,000," MSU physics and astronomy assistant professor Lisa Lapidus said. "Now we can start changing the models - we've been trying to solve protein folding for 50 years, and now we're advancing our fundamental understanding of what unfolded proteins do before they fold."
The findings were published online by the science journal Proceedings of the National Academy of Sciences. Lapidus was joined in the research by University of Zurich Institute of Physical Chemistry researcher Steven Waldauer, whose recent MSU doctoral dissertation formed the basis of the study, and University of California, Davis, scientist Olgica Bakajin.
Proteins, which do most of the work in the body's cells, are chain molecules composed of amino acids. The order in which the amino acids are assembled was charted by the Human Genome Project, but the function of the protein depends on its shape, and how a protein folds is not yet understood. Much of the process is random and diffusive, like sugar moving through an unstirred cup of coffee.
Most proteins can fold in milliseconds, although there are so many possible combinations that left to chance it's physically impossible, scientists agree. So they speculate that there must be built-in folding pathways - but those remain unproved. Now physics is helping make sense of biology, posting a lower speed limit for proteins as they spontaneously assemble into their lowest-energy, so-called natural state - like a relaxed spring.
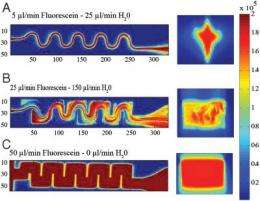
"In order to measure how quickly this random, unfolded state changes confirmations, we had to design an entirely new apparatus as well as design and fabricate a microfluidic chip capable of observing proteins within a fraction of a millisecond after being allowed to refold," Waldauer explained. Two lasers were employed to observe the formation of the immunoglobulin proteins.
"We found that the nature of the unfolded state is far from intuitive and that a protein will change from one random conformation to another much more slowly than previously thought," he said.
Scientists know that errors can occur in folding, and these are associated with a variety of diseases including Alzheimer's, ALS, cystic fibrosis and diabetes. Lapidus and colleagues speculate that the rate of the process could influence the outcome. Proteins that wiggle more rapidly, for example, may be more prone to sticking together and causing plaques such as those in Alzheimer's. The team's discovery may lead to new therapeutic strategies for this class of diseases.
"I believe this measurement of intramolecular diffusion is something that will be crucial for any subsequent studies of protein folding or mis-folding," Lapidus said.
Provided by Michigan State University