Strategy to Quantify, Purify Surface Proteins Also Shows Effects on Protein Translocation
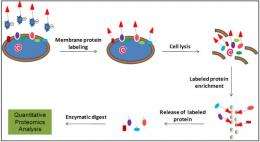
(PhysOrg.com) -- It's always good when you can get two discoveries for the price of one. A strategy developed by scientists at Pacific Northwest National Laboratory to quantify and purify proteins on the surface membranes of cells has also revealed other proteins that have potentially novel roles in cell substrates.
Even more important, the researchers also found that deletion of a type II secretion protein had minimal effects on total protein expression, but significant effects on protein translocation to the cell membrane. Their results will appear in the Journal of Proteome Research.
Surface membrane proteins are essential for maintaining normal biological functions in cells, and often are the "first responders" to environmental stimuli. Despite their biological significance, membrane proteins can be low in abundance and insoluble, making them challenging to quantify and purify. Developing a strategy that can probe changes in membrane protein abundance will improve the understanding of overall biological cellular functions.
The PNNL team met this challenge by first enriching surface membrane proteins expressed by Shewanella oneidensis MR-1 using a membrane-impermeable chemical probe, which allowed labeling of the surface exposed peptides. By linking this method with post-digestion stable isotope labeling, the surface proteins can be quantified. The team identified about 400 proteins, of which 79% were predicted to be localized in the membrane. The successful determination of membrane protein abundance change caused by genetic deletion of one of their translocation pathways further demonstrated the specificity and sensitivity of this strategy in quantifying the membrane proteome abundance.
This work was supported by the U.S. Department of Energy Office of Biological and Environmental Research's (DOE-BER's) Genomics Science Program.
More information: Zhang H, RN Brown, W Qian, ME Monroe, SO Purvine, RJ Moore, MA Gritsenko, L Shi, MF Romine, JK Fredrickson, L Paša-Tolic, RD Smith, and MS Lipton. 2010. "Quantitative analysis of cell surface membrane proteins using membrane-impermeable chemical probe coupled with 18O labeling." Journal of Proteome Research published online April 9, 2010, doi:10.1021/pr9009113
Provided by Pacific Northwest National Laboratory



















