Scientists determine 3D structure of proteins in living cells for the first time
(PhysOrg.com) -- A University of Glasgow scientist was part of a team of researchers which has, for the first time, been able to determine the three-dimensional structure of protein in living cells.
The discovery, published in the latest edition of Nature, means scientists can now prove correct previous assumptions about the structure of proteins and how they change due to mutations and interactions with each other, as well as helping to find ways of correcting damage.
From the 1950s until now, scientists have only been able to closely examine the structures of proteins in their extracted and purified form (in vitro) but these conditions are very different from those inside living cells (in vivo).
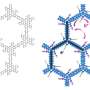
Using a nuclear magnetic resonance (NMR) spectrometer - a machine that allows the distances between the nuclei of atoms within a molecule to be measured - researchers were able to work out the three dimensional shape of an example protein called TTHA1718 which was being produced in living cells of the bacterium E.coli.
Doctor Brian Smith of the Division of Molecular and Cellular Biology at the University of Glasgow provided expertise that helped the Japanese-based and funded international team, led by Yutaka Ito at the Tokyo Metropolitan University, pursue this particular line of research.
Doctor Smith, a lecturer in biochemistry and cell biology, said: “Most proteins don’t exist in isolation; instead they exist in a very crowded environment inside cells where they interact with other molecules and, critically, a large of class of proteins don’t have a definite three-dimensional structure when you take them out of living cells.
“This new, relatively inexpensive method of using NMR spectroscopy means we can now establish the structure of proteins whilst still in live cells and will tell us much more about how they work, and how they change when mutated.
“Our results open new avenues for investigation of protein structures at atomic resolution and how they change in response to biological events in living environments.
“We’ll now try the technique with other, more interesting proteins, which are unstable when you take them out of cells. Ultimately, it could help us discover whether drugs to correct damaged or mutated proteins are working and find new methods of fixing them.”
Proteins are made up of long chains of amino-acids and play essential roles in all aspects of life from metabolism, through detecting and responding to stimuli, to the way organisms are put together. Mutated proteins are implicated in a whole range of illnesses, from cancer to the neurodegenerative condition Huntington’s Disease.
Dr Smith leads a group at the University of Glasgow which uses a 600MHz NMR spectrometer, with a cryogenically cooled probe, to study the structure and functions of proteins and nucleic acids involved in processes in a variety of systems.
More information: A paper on the research entitled, ‘Protein structure determination in living cells by in-cell NMR spectroscopy’, is published in the journal Nature.
Provided by University of Glasgow