Once suspect protein found to promote DNA repair, prevent cancer
An abundant chromosomal protein that binds to damaged DNA prevents cancer development by enhancing DNA repair, researchers at The University of Texas M. D. Anderson Cancer Center report online this week in the Proceedings of the National Academies of Science.
The protein, HMGB1, was previously hypothesized to block DNA repair, senior author Karen Vasquez, Ph.D., associate professor in M. D. Anderson's Department of Carcinogenesis at the Science Park - Research Division in Smithville, Texas.
Identification and repair of DNA damage is the frontline defense against the birth and reproduction of mutant cells that cause cancer and other illnesses.
Pinpointing HMGB1's role in repair raises a fundamental question about drugs under development to block the protein, Vasquez said. The protein also plays a role in inflammation, so it's being targeted in drugs under development for rheumatoid arthritis and sepsis.
"Arthritis therapy involves long-term treatment," Vasquez said. "Our findings suggest that depleting this protein may leave patients more vulnerable to developing cancer."
Long known to attach to sites of damaged DNA, the protein was suspected of preventing repair. "That did not make sense to us, because HMGB1 is a chromosomal protein that's so abundant that it would be hard to imagine cell repair happening at all if that were the case," Vasquez said.
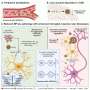
In a series of experiments reported in the paper, Vasquez and first author Sabine Lange, a doctoral candidate in the Graduate School of Biomedical Sciences, tracked the protein's impact on all three steps of DNA restoration: access to damage, repair and repackaging of the original structure, a combination of DNA and histone proteins called chromatin.
First, they knocked out the gene mouse embryonic cells and then exposed cells to two types of DNA-damaging agents. One was UV light, the other a chemotherapy called psoralen that's activated by exposure to darker, low frequency light known as UVA. In both cases, the cells survived at a steeply lower rate after DNA damage than did normal cells.
Next they exposed HMGB1 knockout cells and normal cells to psoralen and assessed the rate of genetic mutation. The knockout cells had a mutation frequency more than double that of normal cells, however, there was no effect on the types of mutation that occurred.
Knock out and normal cells were then exposed to UV light and suffered the same amount of damage. However, those with HMGB1 had two to three times the repair as those without. Evidence suggests that HMGB1 works by summoning other DNA repair factors to the damaged site, Vasquez said.
The last step in DNA repair is called chromatin remodeling. DNA does not exist in a linear structure in the chromosome, but wraps around specialized histone proteins. This chromatin structure permits access to DNA when it is loose, or opened up, and blocks access when it is more tightly wrapped. Presence of HMGB1 resulted in a much higher rate of chromatin assembly in both undamaged and UVC-damaged cells.
Lange and Vasquez hypothesize that HMGB1 normally binds to the entrance and exit of DNA nucleosomes, so is nearby when DNA damage occurs. It then binds to and bends the damaged site at a 90-degree angle, a distortion that may help DNA repair factors recognize and repair the damage. After repair it facilitates restructuring of the chromatin.
Source: University of Texas M. D. Anderson Cancer Center