Genetic 'telepathy'? A bizarre new property of DNA
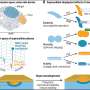
Scientists are reporting evidence that intact, double-stranded DNA has the “amazing” ability to recognize similarities in other DNA strands from a distance. And then like friends with similar interests, the bits of genetic material hangout or congregate together. The recognition — of similar sequences in DNA’s chemical subunits — occurs in a way once regarded as impossible, the researchers suggest in a study scheduled for the Jan. 31 issue of ACS’ Journal of Physical Chemistry B.
Geoff S. Baldwin, Sergey Leikin, John M. Seddon, and Alexei A. Kornyshev and colleagues say the homology recognition between sequences of several hundred nucleotides occurs without physical contact or presence of proteins, factors once regarded as essential for the phenomenon.
This recognition may help increase the accuracy and efficiency of the homologous recombination of genes — a process responsible for DNA repair, evolution, and genetic diversity. The new findings thus may shed light on ways to avoid recombination errors, which underpin cancer, aging, and other health problems.
In the study, scientists observed the behavior of fluorescently tagged DNA strands placed in water that contained no proteins or other material that could interfere with the experiment. Strands with identical nucleotide sequences were about twice as likely to gather together as DNA strands with different sequences.
“Amazingly, the forces responsible for the sequence recognition can reach across more than one nanometer of water separating the surfaces of the nearest neighbor DNA,” said the authors.
Source: ACS