Gene signature spells poor outcome
Other than visually inspecting the disease, doctors have no genetic blueprint to classify melanomas, a lethal form of skin cancer. Tumors generally are ranked by how deeply the growth has invaded underlying skin tissue. The deeper it burrows into the skin, the more lethal the cancer, but some patients defy the odds and survive with thick tumors or die from thin ones.
“Two melanoma patients with cancers of the same invasion depth and appearance under the microscope can have completely different outcomes,” says Rhoda Alani, M.D., associate professor of oncology, dermatology and molecular biology and genetics at Hopkins’ Kimmel Cancer Center.
Alani says the way genes turn their protein-manufacturing machinery on and off in each cancer may help create a signature that can be used to identify tumors that are more prone to kill. These so-called expression patterns can be different from one stage of cancer to the next.
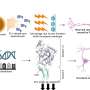
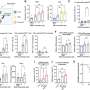
Her research team charted the level of gene expression in melanoma cell lines. Three of the lines mimic the least aggressive type, which grows along the uppermost surface of the skin, called radial growth phase. Four of the cell lines are typical of so-called “vertical growth phase” cancers, which invade inner skin layers, and another three represent the most lethal form -metastatic melanomas.
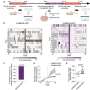
Two vertical growth phase cell lines had gene expression patterns similar to radial growth cancers, indicating that these cells were less aggressive, according to the scientists. The remaining two vertical growth cell lines contained patterns in 18 genes that paralleled metastatic cancer cell lines, the most aggressive form. Alani and her colleagues believe that within this group of 18 genes is a signature for aggressive melanomas.
Many of the genes described in the Hopkins report, published online on July 4 in PLoS One, were previously identified as associated with aggressive cancers by scientists at Johns Hopkins and elsewhere, but Alani says her study brings them all together for melanoma and links them to an aggressive profile.
Alani’s team is validating these results in human tissue samples and evaluating gene correlations with patient outcomes. Funding for the study was provided by the National Cancer Institute. With further study, the genes could be used in tests that predict a patient’s prognosis and as targets for tailored therapies, she says.
Source: Johns Hopkins Medical Institutions