Chemical probes beat antibodies at own game
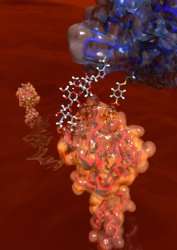
A new way of detecting biological structures could help in the fight against disease. The new method, developed by scientists at Oxford University, uses chemistry to assemble proteins into ‘protein probes’ that can be sent into the body to, for instance, detect inflammation and disease in the brain.
The human body’s immune system uses its own ‘protein probes’ – antibodies – to seek out foreign objects such as bacteria and viruses. These antibodies are proteins shaped to ‘fit’ around parts of a target structure rather like a hand gripping a door handle. Scientists managed to replicate this process for the first time in 1975 with the creation of monoclonal antibodies now widely used in vaccines and biotechnology.
‘We think of antibodies as natural but the ones we use for detection are not always that great at binding to certain structures and in some ways the binding of antibodies to their targets is pretty unrepresentative of other protein-to-protein interactions,’ said Professor Ben Davis of Oxford’s Department of Chemistry who led the work, ‘our protein probes, created using chemical rather than biological techniques, are sometimes better at binding with targets than antibodies because they can mimic natural protein partners more closely.’ By attaching themselves to binding proteins on target cells, such as chronically inflamed brain cells, the probes stain the tissue and this staining can then be detected with a microscope or other methods.
A report on the research, entitled ‘Expanding the diversity of chemical protein modification allows post-translational mimicry’, has just been published in the journal Nature.
It details how Oxford scientists used chemistry to control the modification of proteins from simple chains of amino acids to complex structures similar to those seen in nature. By combining these structures with ready-made protein scaffolds the research team were then able to produce and test protein probes designed to target specific biological structures. In some cases they found that their protein probes were better at binding with some protein partners than monoclonal antibodies.
‘The strategy behind these probes could be useful for making synthetic proteins in many areas of medicine and science’ said Professor Davis ‘but the real goal behind making these modified protein structures using chemistry is that we hope it could enable us to find clues as to how we have ended up with such complex life forms from surprisingly few genes.’
Source: University of Oxford